About the database
Circadian rhythms represent a fundamental regulatory system that coordinates numerous physiological and behavioral processes and are also closely ralated to diseases.
Recent advances in research on the circadian rhythm of brain regions, especially the suprachiasmatic nucleus (SCN), have elucidated gene expression patterns and their functional roles across distinct anatomical subdivisions.
However, understanding of circadian rhythms in other brain regions or across the entire brain remain limited.
Over the past two decades, tissue clearing technique has significantly advanced as a powerful tool, allowing biological tissues to become transparent while preserving structural integrity1,2,3,4.
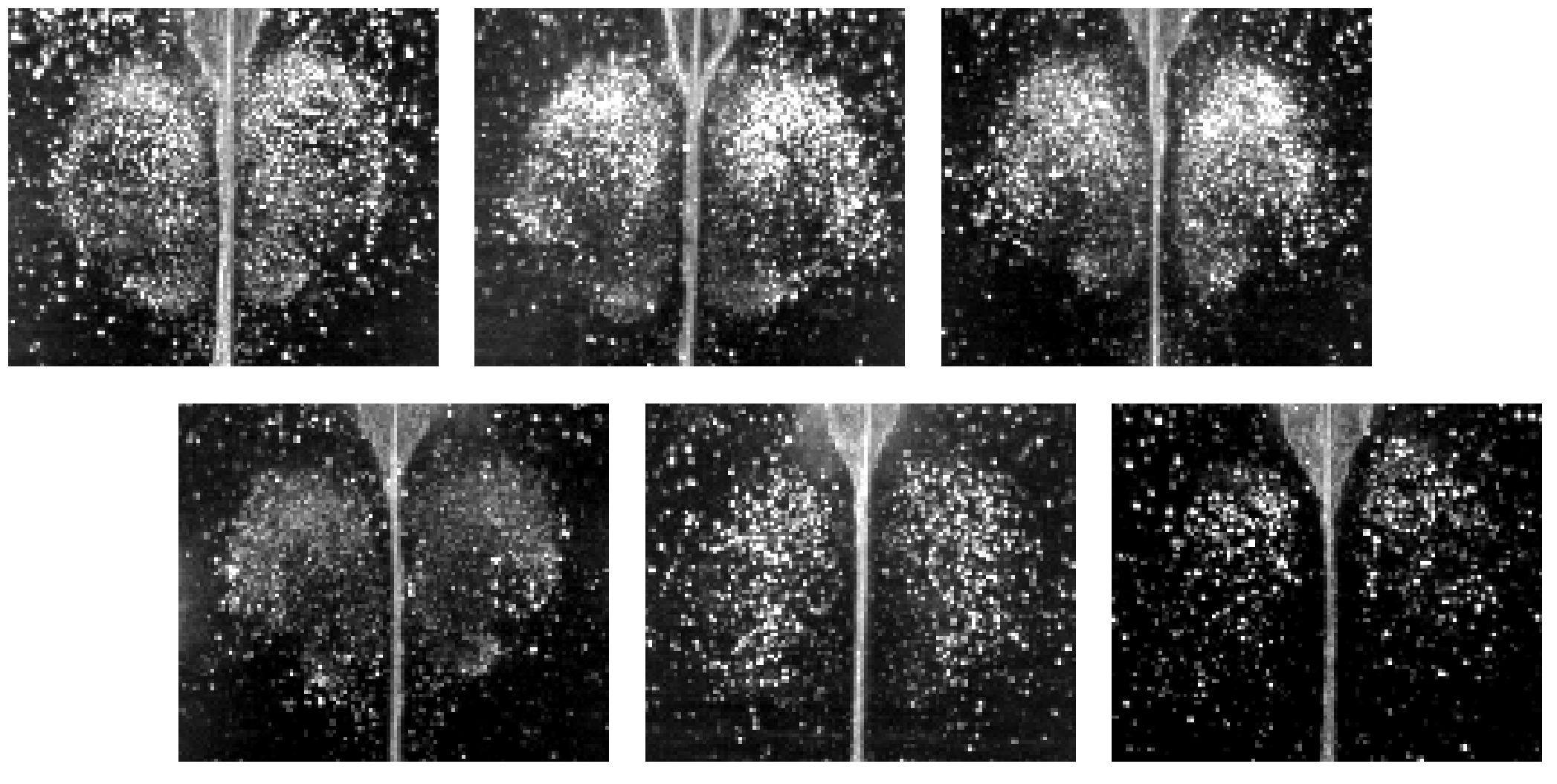
In 2023-2025, we have investigated the circadian rhythm of c-Fos expression over a two-day period in the mouse whole brain under constant darkness (DD) conditions by using Clear, Unobstructed Brain/Body Imaging Cocktails and Computational Analysis (CUBIC) including whole-brain immunostaining4.
Our results demonstrated that substantial brain regions exhibit significant rhythmicity and most regions peak during subjective night.
We registered these data in an open-access database, allowing researchers worldwide to access and utilize the results.
Researchers can refer to the information of circadian changes in counts of c-Fos positive cells, normalized images and the results of detailed rhythmicity analysis at a voxel scale of 20 µm.
The abbreviations for regions are based on the Allen Brain Atlas mouse atlas5.
These data are expected to be integrated with other omics datasets.
We will update the database to obtain more conprehensive understanding of circadian rhythms in the brain.
References
1. Dodt, Hans-Ulrich, Ulrich Leischner, Anja Schierloh, Nina Jährling, Christoph Peter Mauch, Katrin Deininger, Jan Michael Deussing, Matthias Eder, Walter Zieglgänsberger, and Klaus Becker. 2007. “Ultramicroscopy: Three-Dimensional Visualization of Neuronal Networks in the Whole Mouse Brain.” Nature Methods 4 (4): 331–36.
2. Chung, Kwanghun, Jenelle Wallace, Sung-Yon Kim, Sandhiya Kalyanasundaram, Aaron S. Andalman, Thomas J. Davidson, Julie J. Mirzabekov, et al. 2013. “Structural and Molecular Interrogation of Intact Biological Systems.” Nature 497 (7449): 332–37.
3. Renier, Nicolas, Zhuhao Wu, David J. Simon, Jing Yang, Pablo Ariel, and Marc Tessier-Lavigne. 2014. “IDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging.” Cell 159 (4): 896–910.
4. Susaki, Etsuo A., Kazuki Tainaka, Dimitri Perrin, Fumiaki Kishino, Takehiro Tawara, Tomonobu M. Watanabe, Chihiro Yokoyama, et al. 2014. “Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis.” Cell 157 (3): 726–39.
5. Wang, Quanxin, Song-Lin Ding, Yang Li, Josh Royall, David Feng, Phil Lesnar, Nile Graddis, et al. 2020. “The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas.” Cell 181 (4): 936-953.e20.